| Viruses |
Above: a picture of a virus of the bacteriophage type. Click on the image to
enlarge and see the image below for a labeled version. It may look like
something out of science fiction, but these creatures are very real! They are,
however, minute. The one above would be about 200 nanometres tall (a
nanometre is one millionth of a millimetre, so it would take 5000 of them
standing one on top of the other to cover one millimetre in height, and 125 000
to cover an inch!). Not all viruses look like this, in fact they come in a variety of
fantastic shapes and many are much smaller than this one, but very few are a
bit bigger. Most are, however, based on the mathematical shapes known as
the helix and the icosahedron. An icosahedron is a solid 3D shape which has
20 triangular sides. Click here to view the Pov-Ray source code.
enlarge and see the image below for a labeled version. It may look like
something out of science fiction, but these creatures are very real! They are,
however, minute. The one above would be about 200 nanometres tall (a
nanometre is one millionth of a millimetre, so it would take 5000 of them
standing one on top of the other to cover one millimetre in height, and 125 000
to cover an inch!). Not all viruses look like this, in fact they come in a variety of
fantastic shapes and many are much smaller than this one, but very few are a
bit bigger. Most are, however, based on the mathematical shapes known as
the helix and the icosahedron. An icosahedron is a solid 3D shape which has
20 triangular sides. Click here to view the Pov-Ray source code.
Now many biologists do not consider viruses to be living organisms. Whether or not they are organisms as we
know them, they are certainly living in my book. Why? Because they contain genes that replicate over time.
The genes are contained in the icosahedroid head of the virus above. Life is all about information in the form of
complex chemicals such as nucleic acids, such as DNA, and the replication of this information over time. The
debate arises because of the way in which viruses reproduce themselves. Viruses are pirates! They inject their
genes into the cells of other organisms and these genes then take over the host cell and use the cell's
machinery (its mitochondria, endoplasmic reticulum and most importantly the ribosomes) to make more copies
of itself, which then escape from the cell. The cell dies in the process. Thus viruses are pirates that take over
cells. The one above is a pirate of bacteria (it is called a bacteriophage, literally 'bacteria eating'). However,
almost all organisms are dependent on other organisms for their survival. The fact that a virus has to steal
other cells to reproduce itself does not invalidate it being a living creature. Indeed, the concept of an individual
organism is an artificial one - no organism is truly individual! (Well there may be one or two exceptions as we
shall see later).
Viruses are built to be minimal organisms - they are protein shells that carry the minimal number of genes. They
have to be small so that each infected host cell can churn out as many as possible to increase the chances that
the 'hatchlings' will find new host cells. They have no mitochondria and can make no energy of their own (they
are energy parasites) and they have no ribosomes and so cannot make their own proteins. (Some types
contain ribosomes that they carry over from their host cell, but I don't think that these do any work once inside
the virus).
Viruses have ingenious methods of compacting their DNA into as small a volume as possible - they are the
envy of those who are trying to make computer discs that store more information. In fact when the individual
viruses (an individual virus is called a virion) are being assembled minute protein motors wind up their DNA
very tightly and neatly into the protein shell (capsid) and the pressure of the DNA inside may be ten times the
pressure in a bottle of champagne!
The protein shell which you can see in the picture, is called the capsid. The capsid carries the DNA from one
host cell to another. Since the pressure of the tightly packed DNA can be so high, the capsid has to be strong,
and proteins are very strong - if you made a protein large enough to hold in your hand, then it would resemble
very strong plastic.
The bacteriophage shown here has a clever trick: when it lands on a target bacterial cell, the tail tube suddenly
shortens and a sharp needle shoots down through the tough shell of the bacterium and injects the DNA like a
hypodermal syringe!
know them, they are certainly living in my book. Why? Because they contain genes that replicate over time.
The genes are contained in the icosahedroid head of the virus above. Life is all about information in the form of
complex chemicals such as nucleic acids, such as DNA, and the replication of this information over time. The
debate arises because of the way in which viruses reproduce themselves. Viruses are pirates! They inject their
genes into the cells of other organisms and these genes then take over the host cell and use the cell's
machinery (its mitochondria, endoplasmic reticulum and most importantly the ribosomes) to make more copies
of itself, which then escape from the cell. The cell dies in the process. Thus viruses are pirates that take over
cells. The one above is a pirate of bacteria (it is called a bacteriophage, literally 'bacteria eating'). However,
almost all organisms are dependent on other organisms for their survival. The fact that a virus has to steal
other cells to reproduce itself does not invalidate it being a living creature. Indeed, the concept of an individual
organism is an artificial one - no organism is truly individual! (Well there may be one or two exceptions as we
shall see later).
Viruses are built to be minimal organisms - they are protein shells that carry the minimal number of genes. They
have to be small so that each infected host cell can churn out as many as possible to increase the chances that
the 'hatchlings' will find new host cells. They have no mitochondria and can make no energy of their own (they
are energy parasites) and they have no ribosomes and so cannot make their own proteins. (Some types
contain ribosomes that they carry over from their host cell, but I don't think that these do any work once inside
the virus).
Viruses have ingenious methods of compacting their DNA into as small a volume as possible - they are the
envy of those who are trying to make computer discs that store more information. In fact when the individual
viruses (an individual virus is called a virion) are being assembled minute protein motors wind up their DNA
very tightly and neatly into the protein shell (capsid) and the pressure of the DNA inside may be ten times the
pressure in a bottle of champagne!
The protein shell which you can see in the picture, is called the capsid. The capsid carries the DNA from one
host cell to another. Since the pressure of the tightly packed DNA can be so high, the capsid has to be strong,
and proteins are very strong - if you made a protein large enough to hold in your hand, then it would resemble
very strong plastic.
The bacteriophage shown here has a clever trick: when it lands on a target bacterial cell, the tail tube suddenly
shortens and a sharp needle shoots down through the tough shell of the bacterium and injects the DNA like a
hypodermal syringe!
Article updated: 17/1/2015, 2/4/2015, 13/4/2018
You can download the above avi videos from the links below:
Viruses don't just infect bacteria! They also infect animals, plants, algae, protozoa, fungi - in fact probably
every living cell has one or more viruses that parasitise it. Viruses are more-or-less host specific, meaning
that each type can only infect one or a few different types of host cell. There are many countless
bacteriophages multiplying in your own intestines, feeding off the bacteria that grow there, but they are
absolutely incapable of infecting your own cells!
Animal cells are often icosahedral, meaning that they have 20 sides, each a triangle (in a regular icosahedron
these triangles are equilateral triangles, and most icosahedral viruses are modified regular icosahedra), and
also 12 vertices (corners). A standard 20-sided die (d20) is a regular icosahedron. The regular icosahedron
has three different types of symmetry depending which angle it is viewed from. These are shown below:
every living cell has one or more viruses that parasitise it. Viruses are more-or-less host specific, meaning
that each type can only infect one or a few different types of host cell. There are many countless
bacteriophages multiplying in your own intestines, feeding off the bacteria that grow there, but they are
absolutely incapable of infecting your own cells!
Animal cells are often icosahedral, meaning that they have 20 sides, each a triangle (in a regular icosahedron
these triangles are equilateral triangles, and most icosahedral viruses are modified regular icosahedra), and
also 12 vertices (corners). A standard 20-sided die (d20) is a regular icosahedron. The regular icosahedron
has three different types of symmetry depending which angle it is viewed from. These are shown below:
Above a regular icosahedron, left: showing a three-fold axis of symmetry, middle: a five-fold axis of symmetry
and right: a two-fold axis of symmetry.
and right: a two-fold axis of symmetry.
In icosahedral viruses, the icosahedron is a protein shell, called a capsid, made up of protein units called
capsomeres. Each capsomere is a cluster of proteins, with each individual protein being called a protomere. In
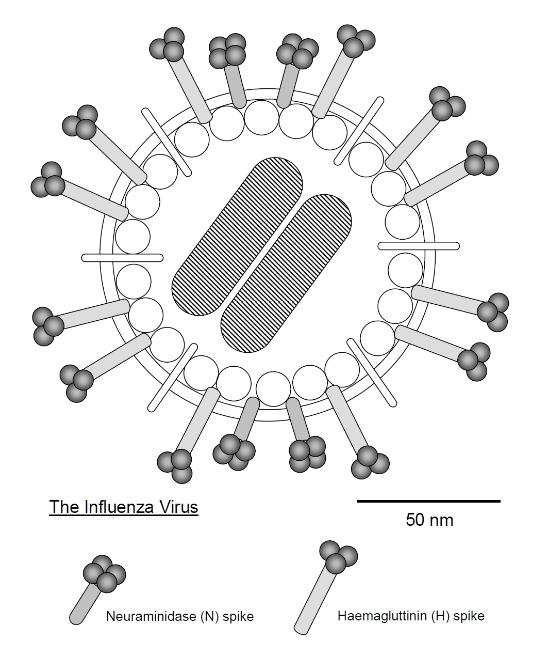
some viruses (e.g. influenza, HIV) the protein capsid in enclosed in a phospholipid membrane (a bilayer
membrane) derived from the host animal cell. Some also contain more than one capsid, with an inner
icosahedron shell inside an outer iscosahedron shell. The capsid (or inner shell when there are more than one)
contains the genetic material of the virus, which may be DNA or RNA (and may be single or double-stranded).
Many animal viruses (such as rabies and ebola), and many plant viruses (such as tobacco mosaic virus, TMV),
and some bacteriophages are filamentous. In this case the filament is a tightly wound helix of protein
capsomeres that form around and enclose the helix of genetic material:
capsomeres. Each capsomere is a cluster of proteins, with each individual protein being called a protomere. In
some viruses (e.g. influenza, HIV) the protein capsid in enclosed in a phospholipid membrane (a bilayer
membrane) derived from the host animal cell. Some also contain more than one capsid, with an inner
icosahedron shell inside an outer iscosahedron shell. The capsid (or inner shell when there are more than one)
contains the genetic material of the virus, which may be DNA or RNA (and may be single or double-stranded).
Many animal viruses (such as rabies and ebola), and many plant viruses (such as tobacco mosaic virus, TMV),
and some bacteriophages are filamentous. In this case the filament is a tightly wound helix of protein
capsomeres that form around and enclose the helix of genetic material:

| Below: Acinetobacter phage 531 with multiple tail disks and disk fibres: |
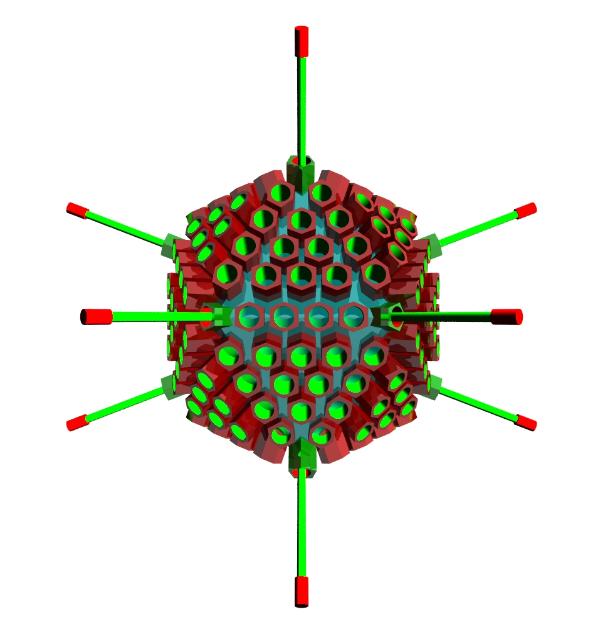
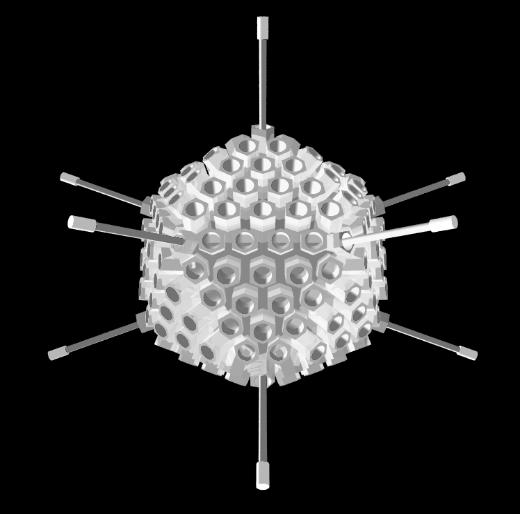
Adenovirus
The Adenoviridae are a group of animal viruses that infect mammals, including humans, birds, reptiles,
amphibians and fish. In humans, most adenoviruses usually cause minor diseases of the upper
respiratory tract, such as the common cold, sore throats, bronchitis and conjunctivitis. However,
complications can occur and some types, such as Ad 14 (adenovirus serotype 14) can be fatal.
The adenovirus is an icosahedral capsid, 90 to 100 nanometres in diameter, made up of six-sided
protein units called hexons and 5-sided pentons forming the vertices (corners). Each vertex also bears
a fibre with a terminal knob. These fibres are involved in attachment to host cells (such as epithelial
cells lining the nose and throat) - the knob sticks to the host cell first, followed by the penton base. The
virus binds to sensors on the cell surface that stimulate the cell to attempt to eat and destroy the virus.
The virus is absorbed by phagocytosis (into what is called a clathrin-coated vesicle, a small fluid-filled
membranous sphere lined by the protein clathrin). The vesicle delivers the virus to an endosome
(essentially the 'stomach' of the cell) for destruction. However, the acidity of the endosome triggers a
reaction in the virus capsid and the pentons affect the virus' escape from the endosome into the cell
cytoplasm. Once free inside the cell, the virus hitches a lift on the cell's own transport system and is
carried along protein tubules (microtubules) in monorail fashion to the cell nucleus (the cell's command
centre). The virus binds to proteins (the nuclear pore complex) that guard the nuclear pores and
control access to and from the nucleus. The virus capsid again deceives the cell with false signals and
releases its DNA (which is a linear double-stranded molecule in adenovirus) which is allowed through
the nuclear pore into the nucleus, the command hub of the cell. Here the viral DNA can utilise the
machinery of the cell, machinery that normally copies and translates the messages inside the cell's own
DNA. Using this machinery the virus replicates itself - it copies its own DNA and translates its encoded
message to produce proteins needed by the virus (in addition to those host cell proteins that the virus
has commandeered). Eventually the viruses induce destruction of the host cell, which bursts, releasing
the newly assembled viruses into the body, where they can affect more cells.
Notice that each triangular face of the virus capsid is made up of 18 hexons and 3 pentons (the 12
edge hexons and the pentons are shared with adjacent faces). An icosahedron has 20 sides, however,
the sharing of some of the capsomeres (hexons and pentons) makes it hard to simply add up how
many there are in the whole virus.
amphibians and fish. In humans, most adenoviruses usually cause minor diseases of the upper
respiratory tract, such as the common cold, sore throats, bronchitis and conjunctivitis. However,
complications can occur and some types, such as Ad 14 (adenovirus serotype 14) can be fatal.
The adenovirus is an icosahedral capsid, 90 to 100 nanometres in diameter, made up of six-sided
protein units called hexons and 5-sided pentons forming the vertices (corners). Each vertex also bears
a fibre with a terminal knob. These fibres are involved in attachment to host cells (such as epithelial
cells lining the nose and throat) - the knob sticks to the host cell first, followed by the penton base. The
virus binds to sensors on the cell surface that stimulate the cell to attempt to eat and destroy the virus.
The virus is absorbed by phagocytosis (into what is called a clathrin-coated vesicle, a small fluid-filled
membranous sphere lined by the protein clathrin). The vesicle delivers the virus to an endosome
(essentially the 'stomach' of the cell) for destruction. However, the acidity of the endosome triggers a
reaction in the virus capsid and the pentons affect the virus' escape from the endosome into the cell
cytoplasm. Once free inside the cell, the virus hitches a lift on the cell's own transport system and is
carried along protein tubules (microtubules) in monorail fashion to the cell nucleus (the cell's command
centre). The virus binds to proteins (the nuclear pore complex) that guard the nuclear pores and
control access to and from the nucleus. The virus capsid again deceives the cell with false signals and
releases its DNA (which is a linear double-stranded molecule in adenovirus) which is allowed through
the nuclear pore into the nucleus, the command hub of the cell. Here the viral DNA can utilise the
machinery of the cell, machinery that normally copies and translates the messages inside the cell's own
DNA. Using this machinery the virus replicates itself - it copies its own DNA and translates its encoded
message to produce proteins needed by the virus (in addition to those host cell proteins that the virus
has commandeered). Eventually the viruses induce destruction of the host cell, which bursts, releasing
the newly assembled viruses into the body, where they can affect more cells.
Notice that each triangular face of the virus capsid is made up of 18 hexons and 3 pentons (the 12
edge hexons and the pentons are shared with adjacent faces). An icosahedron has 20 sides, however,
the sharing of some of the capsomeres (hexons and pentons) makes it hard to simply add up how
many there are in the whole virus.
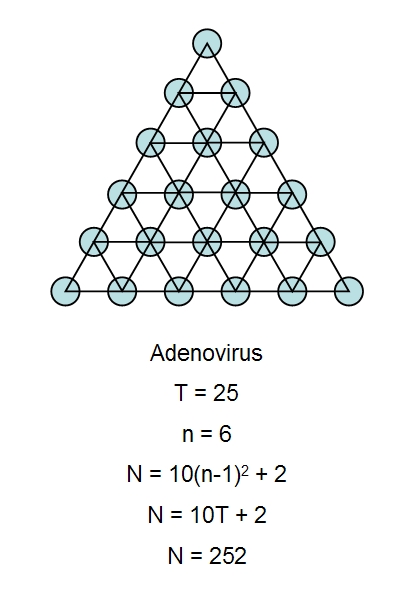
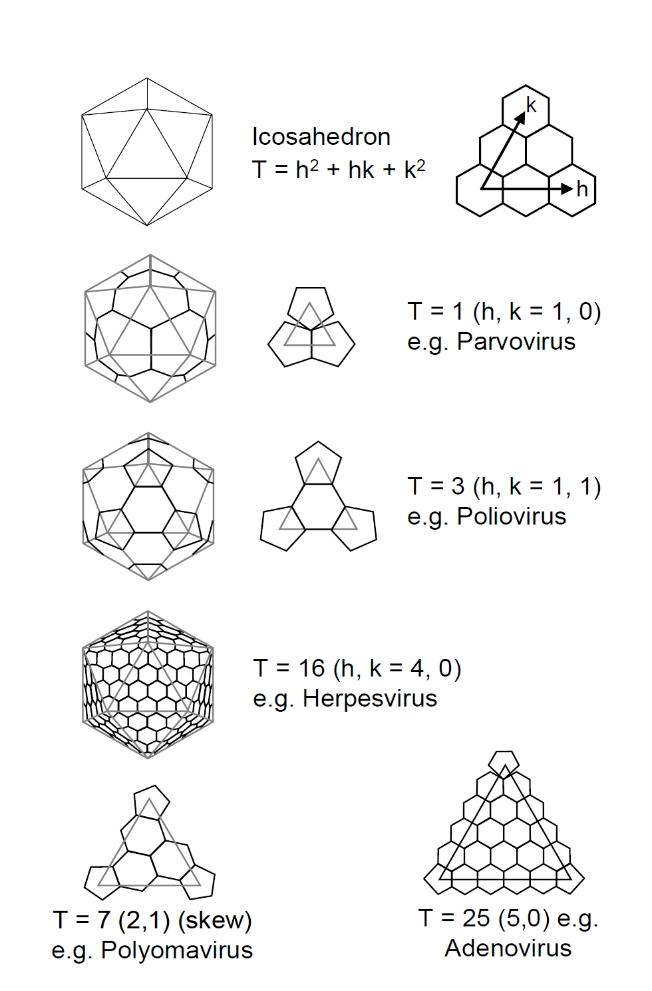
The figure above illustrates the calculation of the total number of capsomeres in a virus with regular
icosahedral symmetry, in this case adenovirus. We have taken a single face of the adenovirus capsid,
which is approximately an equilateral triangle, and we have indicated the position of each capsomere
(hexons and pentons) by blue circles. We have joine dthese up with imaginery small triangles. The total
number of small triangles is the triangulation number, T, which is 25 for adenovirus. Small n designates
the number of capsomeres that make up each adge of the main triangle, 6 in this case. Plugging either
of these values (T or n) into one of the correct formula gives N, the total number of capsomeres in the
icosahedral capsid, N = 252 for adenovirus. In comparison, phage (Phi-X-174) has n = 2, T = 1 and N =
12 capsomeres. The Picorna group viruses, such as poliovirus, consist of a rhombohedron
superimposed upon an icosahedron and require a different formula to calculate N. Each penton is
made up of five protein subunits and each hexon is made up of either six or three protein subunits.
When each hexon contains six protein subunits then we have 60T proteins, however, there are 780
protein subunits in adenovirus (12 x 5 = 60 for the pentons and 240 x 3 = 720 for the hexons).
More geometric arrangements are shown below.
icosahedral symmetry, in this case adenovirus. We have taken a single face of the adenovirus capsid,
which is approximately an equilateral triangle, and we have indicated the position of each capsomere
(hexons and pentons) by blue circles. We have joine dthese up with imaginery small triangles. The total
number of small triangles is the triangulation number, T, which is 25 for adenovirus. Small n designates
the number of capsomeres that make up each adge of the main triangle, 6 in this case. Plugging either
of these values (T or n) into one of the correct formula gives N, the total number of capsomeres in the
icosahedral capsid, N = 252 for adenovirus. In comparison, phage (Phi-X-174) has n = 2, T = 1 and N =
12 capsomeres. The Picorna group viruses, such as poliovirus, consist of a rhombohedron
superimposed upon an icosahedron and require a different formula to calculate N. Each penton is
made up of five protein subunits and each hexon is made up of either six or three protein subunits.
When each hexon contains six protein subunits then we have 60T proteins, however, there are 780
protein subunits in adenovirus (12 x 5 = 60 for the pentons and 240 x 3 = 720 for the hexons).
More geometric arrangements are shown below.
| Download a pdf question on influenza and cell docking |
Tricks of the Trade
We have already seen some of the tricks employed by viruses to infect and hijack cells, but this mode of piracy places
other extreme demands on viruses. Some viruses are much smaller than others, and larger size does mean that more
useful working parts can be incorporated - large viruses like T4 are very efficient at infecting their host. However, all
viruses are minute, since the move from host to host is very uncertain and many virus particles will perish and fail and
never locate or infect a host successfully. Thus, it pays to produce lots of progeny, increasing the odds, but with
limited resources that means keeping viruses small. There is thus a compromise - the virus finds lots of genes useful,
as it has more tools in its toolbox with which to do its job, but space on-board for DNA is very restricted. Viruses have
evolved many ways of packing more into less. First of all, as we have seen, they wind their DNA in tightly, often
reaching enormous internal pressures, which the strong icosahedral protein shells can withstand. Additionally, viruses
have lots of clever tricks of making their DNA very information dense, that is they make a small amount of DNA carry
lots of information. Some of these tricks will surprise those who are familiar only with the way DNA normally functions in
eukaryotes and are as follows:
Two other interesting features of the T4 genome (of uncertain function) are:
We have already seen some of the tricks employed by viruses to infect and hijack cells, but this mode of piracy places
other extreme demands on viruses. Some viruses are much smaller than others, and larger size does mean that more
useful working parts can be incorporated - large viruses like T4 are very efficient at infecting their host. However, all
viruses are minute, since the move from host to host is very uncertain and many virus particles will perish and fail and
never locate or infect a host successfully. Thus, it pays to produce lots of progeny, increasing the odds, but with
limited resources that means keeping viruses small. There is thus a compromise - the virus finds lots of genes useful,
as it has more tools in its toolbox with which to do its job, but space on-board for DNA is very restricted. Viruses have
evolved many ways of packing more into less. First of all, as we have seen, they wind their DNA in tightly, often
reaching enormous internal pressures, which the strong icosahedral protein shells can withstand. Additionally, viruses
have lots of clever tricks of making their DNA very information dense, that is they make a small amount of DNA carry
lots of information. Some of these tricks will surprise those who are familiar only with the way DNA normally functions in
eukaryotes and are as follows:
- Nested genes, or genes within genes. For example, in the T4 genome there at least 5 genes with functional
internal start codons, that is each gene has another, smaller functional gene inside it! RNA polymerase might
start on the outside gene, producing a larger mRNA or sometimes it might start at teh inside gene and produce
a smaller mRNA. At least some of the products of these inner genes have been shown to have distinct and
useful functions. - Closely spaced start codons. At least 5 T4 genes have two start codons, similar to the nested genes, but with
the larger outer gene just having an extra bit tagged on front. In the lambda phage, there is one documented
case where the two products serve distinct functions, with the larger protein have only two extra amino acids at
the start but with one protein functioning as a pore for lysozyme to reach the peptidoglycan and degrade it,
whilst the other protein delays pore formation (and so regulates the function of the first). - Nested genes may have different reading frames. Recall that every three consecutive bases make one codon
which codes for one amino acid. One codon follows another, every 3 bases, on a DNA molecule. Depending
where we start reading the DNA we get very different sequences that encode very different proteins. Inner
genes might be read 'out-of-sink' with their outer container gene in this fashion, and they may be read in the
same or the opposite direction. Some viruses (though not T4) have a programmed frameshift, in which the
genome can be re-read but with the sequence moved forwards or backwards by one base, resulting in very
different codons. - Overlapping genes. frequently the stop codon of one gene overlaps the start codon of the following gene.
regulatory regions of DNA may also overlap coding regions. - Few noncoding regions. Noncoding regions of DNA are very abundant in eukaryotes. Some of this DNA
performs other useful functions, such as switching genes on or off, some may be junk DNA or parasitic DNA. In
T4 only 5.3% of the genome (9 kb) is non-coding. - Resistance to host cell nucleases. Bacteria and other cells have evolved nuclease enzymes that recognise
foreign DNA, including viral DNA, and destroy it. T4 has modified its DNA, it has replaced all the cytosine (C
base) with a modified form, hydroxymethylcytosine (HMC). However, Escherichia coli has nucleases that can
recognise this viral base, but T4 counters this by another modification, it glucosylates (adds a glucose residue
to) the HMC, forming glucosyl-HMC. T4 genome also has an unusual high proportion of A and T bases
(65.5% of the bases with 34.5% G+C) instead of the 50% expected from the law of averages. This gives the
phage DNA a DNA similar in structure to D-DNA (that is DNA made up entirely of A-T base pairs) that is it has
only 8 base pairs per turn of the double helix (instead of the usual 10) and a wider, shallower major groove and
a deeper, narrower minor groove. These spiral grooves of DNA are used by many enzymes to grab hold of DNA
and so changes in these grooves may also affect the binding of host DNA-recognising enzymes. Host cells
contain exonucleases that degrade foreign DNA at its free ends, to protect against this a linear dsDNA phage
genome may join its ends together or circularise, forming circular dsDNA upon entering the host cell
cytoplasm. Lambda phage is known to do this.
Two other interesting features of the T4 genome (of uncertain function) are:
- Translational bypassing, e.g. in T4 gene 60 a 50 base mRNA segment is bypassed by the ribosome. Is this
segment always bypassed or does this gene produce two different proteins depending whether or not the
bypass occurs? - Introns occur in at least three T4 genes. These are sequences of DNA inside a gene which are removed later
on in transcription. These are normally associated with eukaryotes. In T4 the introns are self-splicing, that is
they have enzymatic properties and can remove themselves from the gene whose code they interrupt. Whether
these introns serve a useful function in generating alternative sequences or whether they are just parasitic
genes hitching a ride is not clear (they will infect the DNA of intron-less virus DNA in the same host).
The most complex viruses include the vaccinia pox virus and T4 bacteriophage. T4 has a genome consisting of
circular double-stranded DNA with 168903 bp (base-pairs) and about 289 protein-encoding genes, 8 tRNA genes
(see ribosomes) and at least 2 genes that encode small RNA molecules of unknown function.
A more detailed sequence of events in T4 DNA injection into the host bacterial cell is given below (for this account it
is necessary to understand the basic structure of bacterial cell envelopes):
Once inside the host cell, the DNA has to compete with the host DNA for the transcription and translation machinery
of the cell. The promoters (control regions of the DNA which control transcription) of T4 are stronger than those of
the host and are preferentially transcribed into mRNA. Gene transcription is divided into three phases, early,
middle and late transcription (a common virus strategy) with the early genes being transcribed first during the
infection cycle. Host gene expression is also shut down rapidly, leaving all the cell's machinery at the virus' disposal.
A T4 protein (gpAlt) enters the host cell along with the DNA and this virus enzymically alters the host RNA
polymerase, the enzyme responsible for transcribing DNA into mRNA for protein synthesis. (Specifically it ADP-
ribosylates the polymerase enzyme). This modified polymerase preferentially recognises the T4 promoters - the
macinery of the cell is thus rapidly commandeered! Two of the early genes transcribed code for the proteins ModA
and ModB which switch-off host promoters, shutting down host gene expression completely. Eventually, these
proteins turn-off the early promoters, since these have done their job of rapidly securing the cell for viral purposes.
Another T4 protein, AsiA modifies the RNA polymerase further, priming it to transcribe the middle genes. The late
genes include those for head, tail and tail fibre synthesis and assembly (this requires 54 genes, of which 5 encode
catalyst enzymes, the other 49 structural components; of these 54, 24 are needed for the head, 22 for the tail and 7
for the tail fibers (including one to attach the tail fibers)).
The DNA must be packaged into the head. Many viruses with circular genomes of double-stranded DNA replicate
their DNA by rolling-circle replication, in which a template strand rotates, churning out a chain of many copies of
itself, called a concatemer. This DNA is wound tightly into the head by molecular motors (the neck of the T4 phage,
the disc-like structure between the head and collar is thought to rotate as DNA as wound tightly into the head).
When the head is full, the concatemer is cut. Normally 103% of a genome gets incorporated into the head, that is
one complete copy of the DNA plus a little bit at the ends, called terminal redundancy. The concatemer then
continues feeding into another phage. Sometimes defects occur and the head is too small to contain the whole
genome, in which case the virus usually can not complete the life-cycle by itself, but must co-infect a host with
another intact virus (which presumably it can do if both infect within 4 minutes of each other). Defective large heads
contain spare DNA. Eventually some 100 to 150 new T4 phages are assembled. Another T4 lysozyme then
degrades the host cell wall, causing the cell to burst or lyse and release the new virus particles so that they can
infect new hosts! T4 has another clever trick to avoid excessive competition with other phages - lysis inhibition.
The virus inside the host can sense the presence of phages outside (perhaps as they attempt to infect the cell) and
if there are too many, then they will delay lysis, waiting instead to be released under less competitive conditions!
circular double-stranded DNA with 168903 bp (base-pairs) and about 289 protein-encoding genes, 8 tRNA genes
(see ribosomes) and at least 2 genes that encode small RNA molecules of unknown function.
A more detailed sequence of events in T4 DNA injection into the host bacterial cell is given below (for this account it
is necessary to understand the basic structure of bacterial cell envelopes):
- At least 3 of the 6 tail fibers must bind to the host. They bind to a glucose residue on the lipopolysaccharide
(LPS) outer core. (The outer membrane of Gram-negative bacteria is composed in large part of LPS which
contains chains of sugars bonded to one-another). - The baseplate changes configuration from a hexagon to a star-shape, deploying the six tail pins. The short
tail fibers, previously contained inside the baseplate, are deployed and these bind to a heptose sugar residue
on the LPS inner core, forming a second and more stable adhesion to the host cell. - Simultaneous to the baseplate change is the contraction of the tail-sheath, extruding the inner tail tube which
punctures the outer membrane. The energy for the contraction was stored as potential mechanical energy
when the phage was assembled - it is a primed or cocked mechanical needle. - The terminal needle detaches and a protein, gp5, in the baseplate is a lysozyme enzyme which degrades the
peptidoglycan layer. - The DNA exits through the tail tube tip and enters the periplasm of the host (this involves a host
phosopholipid in the inner membrane, called phosphatidylglycerol). The electric potential across the inner
membrane (which as in all living cells acts as an electric capacitor and stores electric charge) is essential for
DNA entry across the inner membrane and into the cytoplasm. - Within 4 minutes of DNA entry (infection) further infecting phage DNA gets trapped in the periplasm and is
degraded by nuclease enzymes. This involves the imm T4 gene and prevents superinfection and
competition for the cell's resources with other phages - the first phage in gets the prize!
Once inside the host cell, the DNA has to compete with the host DNA for the transcription and translation machinery
of the cell. The promoters (control regions of the DNA which control transcription) of T4 are stronger than those of
the host and are preferentially transcribed into mRNA. Gene transcription is divided into three phases, early,
middle and late transcription (a common virus strategy) with the early genes being transcribed first during the
infection cycle. Host gene expression is also shut down rapidly, leaving all the cell's machinery at the virus' disposal.
A T4 protein (gpAlt) enters the host cell along with the DNA and this virus enzymically alters the host RNA
polymerase, the enzyme responsible for transcribing DNA into mRNA for protein synthesis. (Specifically it ADP-
ribosylates the polymerase enzyme). This modified polymerase preferentially recognises the T4 promoters - the
macinery of the cell is thus rapidly commandeered! Two of the early genes transcribed code for the proteins ModA
and ModB which switch-off host promoters, shutting down host gene expression completely. Eventually, these
proteins turn-off the early promoters, since these have done their job of rapidly securing the cell for viral purposes.
Another T4 protein, AsiA modifies the RNA polymerase further, priming it to transcribe the middle genes. The late
genes include those for head, tail and tail fibre synthesis and assembly (this requires 54 genes, of which 5 encode
catalyst enzymes, the other 49 structural components; of these 54, 24 are needed for the head, 22 for the tail and 7
for the tail fibers (including one to attach the tail fibers)).
The DNA must be packaged into the head. Many viruses with circular genomes of double-stranded DNA replicate
their DNA by rolling-circle replication, in which a template strand rotates, churning out a chain of many copies of
itself, called a concatemer. This DNA is wound tightly into the head by molecular motors (the neck of the T4 phage,
the disc-like structure between the head and collar is thought to rotate as DNA as wound tightly into the head).
When the head is full, the concatemer is cut. Normally 103% of a genome gets incorporated into the head, that is
one complete copy of the DNA plus a little bit at the ends, called terminal redundancy. The concatemer then
continues feeding into another phage. Sometimes defects occur and the head is too small to contain the whole
genome, in which case the virus usually can not complete the life-cycle by itself, but must co-infect a host with
another intact virus (which presumably it can do if both infect within 4 minutes of each other). Defective large heads
contain spare DNA. Eventually some 100 to 150 new T4 phages are assembled. Another T4 lysozyme then
degrades the host cell wall, causing the cell to burst or lyse and release the new virus particles so that they can
infect new hosts! T4 has another clever trick to avoid excessive competition with other phages - lysis inhibition.
The virus inside the host can sense the presence of phages outside (perhaps as they attempt to infect the cell) and
if there are too many, then they will delay lysis, waiting instead to be released under less competitive conditions!
Rabies is caused by a rhabdovirus (above, click images to enlarge). Rabies
apparently infects every mammal species and in addition to domestic cats and dogs it
is most often contracted from skunks, raccoons, bats and foxes. Infection occurs when
an infected or rhabid animal bites or licks an open wound. Occasionally it has also
been contracted by inhalation of virus-containing aerosol in bat caves. It has also
been contracted from corneal transplants (from donors who died of other causes).
Once inside the body the virus infects peripheral nerve cells and moves along them to
the central nervous system. Once inside the brain it triggers an inflammatory response
(encephalitis) which is almost always fatal. The death rate of those contracting the
systems is almost 100%. No more than a handful of people are reported to have
survived having the symptoms of rabies. However, an effective vaccine is available
(consisting of the dead and inactivated virus) which confers immunity to those yet to
be infected and is also effective if administered rapidly after being bitten. Symptoms of
rabies appear 2-16 weeks after initial infection and are: fatigue, loss of appetite, fever
and often a tingling or burning sensation at the site of the wound and so-called
'hydrophobia' occurs in 50% of cases, in which swallowing causes painful spasms in
throat and chest muscles.
apparently infects every mammal species and in addition to domestic cats and dogs it
is most often contracted from skunks, raccoons, bats and foxes. Infection occurs when
an infected or rhabid animal bites or licks an open wound. Occasionally it has also
been contracted by inhalation of virus-containing aerosol in bat caves. It has also
been contracted from corneal transplants (from donors who died of other causes).
Once inside the body the virus infects peripheral nerve cells and moves along them to
the central nervous system. Once inside the brain it triggers an inflammatory response
(encephalitis) which is almost always fatal. The death rate of those contracting the
systems is almost 100%. No more than a handful of people are reported to have
survived having the symptoms of rabies. However, an effective vaccine is available
(consisting of the dead and inactivated virus) which confers immunity to those yet to
be infected and is also effective if administered rapidly after being bitten. Symptoms of
rabies appear 2-16 weeks after initial infection and are: fatigue, loss of appetite, fever
and often a tingling or burning sensation at the site of the wound and so-called
'hydrophobia' occurs in 50% of cases, in which swallowing causes painful spasms in
throat and chest muscles.
Rhabdoviruses are rod-shaped or bullet-shaped filamentous viruses. They are grouped together with the filoviruses (such
as ebolavirus) and the paramyxoviruses (such as measles, mumps and Newcastle disease viruses). These viruses have
genomes of negative-sense single-stranded RNA (ss(-)RNA). Negative-sense means that the RNA is complementary to
the actual coding RNA and so must be copied before it can be translated into proteins by the host cell's ribosomes. Naked
rabies virus genomic RNA is not infectious, as it requires a copy of the viral-encoded polymerase (RNA-dependent RNA
polymerase) to read it and make the complementary RNA strand (synthesising RNA from an RNA template is not a normal
process in cells). This is in contrast to poliovirus which has a genome of ss(+)RNA which acts as a messenger RNA and so
can be read directly by the ribosomes and translated - naked poliovirus RNA is infectious when injected into cells (the
poliovirus capsid is still needed to infect the specific target cells in nature).
The rabies virus is filamentous, and typical of filamentous viruses it's chief component is the helical ribonucleoprotein core
(RNA bound to stabilising nucleoprotein and wound into a helix). RNA is an unstable molecule, but binding it to proteins in
this way protects it and packages it. This is covered by a cylinder of M-protein (although in vesicular stomatitis virus (VSV) it
has been suggested that the M-protein forms the axial core instead). Wherever it is situated it triggers coiling of the
ribonucleoprotein. This viral core is sheathed in phospholipid bilayer membrane - unit membrane derived from the host cell
which is modified to contain the viral glycoprotein spikes.
The rabies genome is about 12 kb long and contains five genes as follows:
Rabies virus genome (~12kb, 5 genes) ss(–)RNA:
3’–(LDR)–N–P–M–G–L–(TLR)-5’
LDR: leader sequence (about 50 nucleotides) at the 3' end
TLR: trailer sequence at the 5' end
Compare this to the ebolavirus genome, which is larger and contains 7 genes:
Ebola virus genome (~19kb, 7 genes) ss(–)RNA:
3’–(LDR)–NP–VP35–VP40–GP–VP30–VP24–L–(TLR)-5’
Note that the equivalent proteins found in rabies virus are all present in addition to VP30 and VP24. The phosphoprotein,
VP35, in addition to being a co-factor for the viral L polymerase and a structural protein (part of the ribonucleoprotein core)
has anti-interferon (IFN) functions. Interferon is a key biochemical involved in triggering the host cell's defences to viral
attack. VP35 inhibits the activation of interferon regulatory factors (IRF-3 and IRF-7). These regulatory factors act as
sensors for viral infection and activate to raise the alarm, they are transcription factors and when activated they move to
the cell nucleus and switch on anti-viral genes, including those that manufacture interferon. By blocking this alarm signal,
ebolavirus can infect cells much more easily. People who recover from infection contain antibodies to NP primarily, but also
the M protein and VP35. Antibodies are produced by the immune system and bind to (and hence inactivate) key pathogen
proteins and other macromolecules called antigens. Presumably, the phosphoprotein of rabies virus has a similar function.
Some 20-90% of those infected die.
Viral Life-Cycles: The Infectious-Cycle of Rabies Virus
Viral Life-Cycles: Lysis and Lysogeny in Phages
The life-cycle exhibited by T4, as described above, is called a lytic cycle. The virus enters the cell and then takes over the
cell's machinery to synthesise more virus units and then viral enzymes degrade the cell wall and the cell suddenly bursts
open (lyses) releasing all of the mature phages. There is an eclipse phase of 20-25 minutes after infection, during which
no mature phage are produced and released, because they are being manufactured and assembled. The average number
of new phage released in lysis is called the burst size and depends on the bacteriophage type.
Another bacteriophage, the lambda bacteriophage (or phage lambda) has a choice and sometimes it undergoes the lytic
cycle, but other times it undergoes the so-called lysogenic life-cycle instead. In lysogeny the viral DNA inserts into the
host cell's chromosome and is maintained in a silent stable state indefinitely, without killing the host and replicating with it. In
this state the phage is called a prophage. Eventually the prophage may switch back to the lytic cycle, removing itself from
the host chromosome and reproducing, assembling phages and bursting the host cell. The lytic cycle is switched on when
the host cell is growing rapidly in nutrient-rich conditions, but if the cell is growing slowly under nutrient-poor conditions then
the lysogenic cycle is switched on. This helps ensure that should lysis occur, there will be plenty of bacterial cells around
for the viral progeny to infect. All this is controlled by a fascinating molecular switch which we may consider in detail, along
with more fascinating facts about phages, in a future article. The bacteriophage M13 is unusual in that mature phages are
constantly produced and liberated from the host cell without killing it.
In animal cells, virus particles are often released by budding rather than by lysis. When this happens, viral membrane
proteins assemble in clusters in the host cell membrane (displacing unwanted host membrane proteins) and then the
assembled viral capsid fuses with this membrane patch which evaginates and buds off from the host cell with a single virus
enclosed. In this way the virus acquires a phospholipid bilayer membrane envelope, containing host-derived lipids and
viral proteins. These are called enveloped viruses. Most animal viruses are enveloped. Naked viruses must exit the cell by
other means, such as by cell lysis.
Virus Assembly
Many viruses are totally self-assembling. The various proteins that they have synthesised bond together spontaneously
to give the final virus product. A source of energy is needed, however, to package DNA into the capsid, this energy (in the
form of ATP from the host cell) powers the molecular motor that winds DNA tightly into the capsid (a process called
encapsidation). In viruses with a complex binary architecture, like T4, the process is, unlike that in icosahedral or
filamentous viruses, not total self-assembly as it requires proteins and enzymes that are not part of the final virus particle.
For example, scaffolding proteins are required for assembly of the phage head and these are later removed.
as ebolavirus) and the paramyxoviruses (such as measles, mumps and Newcastle disease viruses). These viruses have
genomes of negative-sense single-stranded RNA (ss(-)RNA). Negative-sense means that the RNA is complementary to
the actual coding RNA and so must be copied before it can be translated into proteins by the host cell's ribosomes. Naked
rabies virus genomic RNA is not infectious, as it requires a copy of the viral-encoded polymerase (RNA-dependent RNA
polymerase) to read it and make the complementary RNA strand (synthesising RNA from an RNA template is not a normal
process in cells). This is in contrast to poliovirus which has a genome of ss(+)RNA which acts as a messenger RNA and so
can be read directly by the ribosomes and translated - naked poliovirus RNA is infectious when injected into cells (the
poliovirus capsid is still needed to infect the specific target cells in nature).
The rabies virus is filamentous, and typical of filamentous viruses it's chief component is the helical ribonucleoprotein core
(RNA bound to stabilising nucleoprotein and wound into a helix). RNA is an unstable molecule, but binding it to proteins in
this way protects it and packages it. This is covered by a cylinder of M-protein (although in vesicular stomatitis virus (VSV) it
has been suggested that the M-protein forms the axial core instead). Wherever it is situated it triggers coiling of the
ribonucleoprotein. This viral core is sheathed in phospholipid bilayer membrane - unit membrane derived from the host cell
which is modified to contain the viral glycoprotein spikes.
The rabies genome is about 12 kb long and contains five genes as follows:
Rabies virus genome (~12kb, 5 genes) ss(–)RNA:
3’–(LDR)–N–P–M–G–L–(TLR)-5’
LDR: leader sequence (about 50 nucleotides) at the 3' end
- N: nucleoprotein, signals the switch from transcription to regulation and is a structural component, binding to and
packaging the genomic RNA into ribonucleoprotein (RNP);
- P: phosphoprotein;
- M: matrix protein, forms a complex with the RNP just beneath the phospholipid bilayer membrane, induces coiling of
the RNP;
- G: glycoprotein (GP) spikes (involved in host cell recognition, adhesion and penetration) and inserted into the bilayer
phospholipid membrane;
- L: (large protein) viral RNA-dependent RNA polymerase;
- N, P and L: together with RNA form the RNP core;
TLR: trailer sequence at the 5' end
Compare this to the ebolavirus genome, which is larger and contains 7 genes:
Ebola virus genome (~19kb, 7 genes) ss(–)RNA:
3’–(LDR)–NP–VP35–VP40–GP–VP30–VP24–L–(TLR)-5’
- NP: nucleoprotein
- VP35: phosphoprotein
- VP40: matrix protein
- GP: surface spike glycoprotein
- VP30: a minor nucleocapsid component
- VP24: a minor matrix protein
- L: RNA-dependent RNA polymerase
Note that the equivalent proteins found in rabies virus are all present in addition to VP30 and VP24. The phosphoprotein,
VP35, in addition to being a co-factor for the viral L polymerase and a structural protein (part of the ribonucleoprotein core)
has anti-interferon (IFN) functions. Interferon is a key biochemical involved in triggering the host cell's defences to viral
attack. VP35 inhibits the activation of interferon regulatory factors (IRF-3 and IRF-7). These regulatory factors act as
sensors for viral infection and activate to raise the alarm, they are transcription factors and when activated they move to
the cell nucleus and switch on anti-viral genes, including those that manufacture interferon. By blocking this alarm signal,
ebolavirus can infect cells much more easily. People who recover from infection contain antibodies to NP primarily, but also
the M protein and VP35. Antibodies are produced by the immune system and bind to (and hence inactivate) key pathogen
proteins and other macromolecules called antigens. Presumably, the phosphoprotein of rabies virus has a similar function.
Some 20-90% of those infected die.
Viral Life-Cycles: The Infectious-Cycle of Rabies Virus
- Adsorption - the virus glycoprotein (GP) spikes bind to specific receptors on the target cell.
- This triggers receptor-mediated endocytosis (pinocytosis) in which a pit forms in the target cell membrane at the site
of virus adsorption. This pit is lined (on the cytoplasm side) by a cell protein called clathrin, characteristic of this form
of endocytosis. The clathrin facilitates the invagination of the pit into an endocytic vesicle which encloses the
adsorbed virus.
Viral Life-Cycles: Lysis and Lysogeny in Phages
The life-cycle exhibited by T4, as described above, is called a lytic cycle. The virus enters the cell and then takes over the
cell's machinery to synthesise more virus units and then viral enzymes degrade the cell wall and the cell suddenly bursts
open (lyses) releasing all of the mature phages. There is an eclipse phase of 20-25 minutes after infection, during which
no mature phage are produced and released, because they are being manufactured and assembled. The average number
of new phage released in lysis is called the burst size and depends on the bacteriophage type.
Another bacteriophage, the lambda bacteriophage (or phage lambda) has a choice and sometimes it undergoes the lytic
cycle, but other times it undergoes the so-called lysogenic life-cycle instead. In lysogeny the viral DNA inserts into the
host cell's chromosome and is maintained in a silent stable state indefinitely, without killing the host and replicating with it. In
this state the phage is called a prophage. Eventually the prophage may switch back to the lytic cycle, removing itself from
the host chromosome and reproducing, assembling phages and bursting the host cell. The lytic cycle is switched on when
the host cell is growing rapidly in nutrient-rich conditions, but if the cell is growing slowly under nutrient-poor conditions then
the lysogenic cycle is switched on. This helps ensure that should lysis occur, there will be plenty of bacterial cells around
for the viral progeny to infect. All this is controlled by a fascinating molecular switch which we may consider in detail, along
with more fascinating facts about phages, in a future article. The bacteriophage M13 is unusual in that mature phages are
constantly produced and liberated from the host cell without killing it.
In animal cells, virus particles are often released by budding rather than by lysis. When this happens, viral membrane
proteins assemble in clusters in the host cell membrane (displacing unwanted host membrane proteins) and then the
assembled viral capsid fuses with this membrane patch which evaginates and buds off from the host cell with a single virus
enclosed. In this way the virus acquires a phospholipid bilayer membrane envelope, containing host-derived lipids and
viral proteins. These are called enveloped viruses. Most animal viruses are enveloped. Naked viruses must exit the cell by
other means, such as by cell lysis.
Virus Assembly
Many viruses are totally self-assembling. The various proteins that they have synthesised bond together spontaneously
to give the final virus product. A source of energy is needed, however, to package DNA into the capsid, this energy (in the
form of ATP from the host cell) powers the molecular motor that winds DNA tightly into the capsid (a process called
encapsidation). In viruses with a complex binary architecture, like T4, the process is, unlike that in icosahedral or
filamentous viruses, not total self-assembly as it requires proteins and enzymes that are not part of the final virus particle.
For example, scaffolding proteins are required for assembly of the phage head and these are later removed.
Above: a rhabdovirus in section: about 400
copies of the glycoprotein spikes (GP) (in green)
cover the outer surface and anchor in the bilayer
phospholipid membrane (cyan) which covers the
matrix (M, thin outer red layer) which covers the
ribonucleoprotein (RNP) core (RNA bound to
nucleoprotein, N, shown in yellow). The axial core
is shown in red. Length 180 nm, diameter 75 nm.
copies of the glycoprotein spikes (GP) (in green)
cover the outer surface and anchor in the bilayer
phospholipid membrane (cyan) which covers the
matrix (M, thin outer red layer) which covers the
ribonucleoprotein (RNP) core (RNA bound to
nucleoprotein, N, shown in yellow). The axial core
is shown in red. Length 180 nm, diameter 75 nm.
Rhabdoviruses
TMV (Tobacco Mosaic Virus)
Above: TMV, a virus of tobacco plants, is a filamentous RNA virus. Top right: some of the capsid proteins (cyan) have been stripped away
to reveal the RNA helix (yellow). The capsid normally consists of 2130 coat protein molecules (with 16.3 units per turn of the helix) and is
about 300 nm long by 18 nm in diameter (with an inner 4 nm channel). (The model above only shows 480 subunits). The genome is
ss(+)RNA, that is a single-strand of positive-sense RNA, sense meaning that it can be directly transcribed by ribosomes. The RNA is
located at about 6nm from the centre and fits in a helical groove formed by the capsid proteins, with about 3 nucleotides per protein unit.
The proteins stabilise the RNA and the virus can survive temperatures as high 120 C for 30 minutes.
The RNA enters a host cell and the RNA becomes active mRNA and uses the host cell ribosomes to manufacture coat proteins and an
RNA-dependent RNA polymerase (RDRP) which copies the RNA to make more RNA genomes. The protein units (protomers)
self-assemble into discs of 2 layers in a helical spiral, and more protomers add to the end of the rod. A copy of the RNA passes through
the central channel and forms a helix. In this way an infected plant cell creates many copies of the virus which are then able to infect
adjacent cells by moving through the plasmodesmata. The plasmodesmata channels, which connect adjacent cells in plants, are normally
too narrow, but the virus produces a protein called P30 (movement protein) which enlarges the plasmodesmata.
to reveal the RNA helix (yellow). The capsid normally consists of 2130 coat protein molecules (with 16.3 units per turn of the helix) and is
about 300 nm long by 18 nm in diameter (with an inner 4 nm channel). (The model above only shows 480 subunits). The genome is
ss(+)RNA, that is a single-strand of positive-sense RNA, sense meaning that it can be directly transcribed by ribosomes. The RNA is
located at about 6nm from the centre and fits in a helical groove formed by the capsid proteins, with about 3 nucleotides per protein unit.
The proteins stabilise the RNA and the virus can survive temperatures as high 120 C for 30 minutes.
The RNA enters a host cell and the RNA becomes active mRNA and uses the host cell ribosomes to manufacture coat proteins and an
RNA-dependent RNA polymerase (RDRP) which copies the RNA to make more RNA genomes. The protein units (protomers)
self-assemble into discs of 2 layers in a helical spiral, and more protomers add to the end of the rod. A copy of the RNA passes through
the central channel and forms a helix. In this way an infected plant cell creates many copies of the virus which are then able to infect
adjacent cells by moving through the plasmodesmata. The plasmodesmata channels, which connect adjacent cells in plants, are normally
too narrow, but the virus produces a protein called P30 (movement protein) which enlarges the plasmodesmata.

In the lambda bacteriophage, the linear dsDNA chromosome circularises on entering the host and initially
replicates by theta-replication, a form of bidirectional replication similar to that in bacteria like Escherichia
coli (see growth in bacteria). These initial copies ensure adequate production of viral products to initiate the
infection cycle, but later on, when assembling mature phages and packaging DNA into the phage heads it
resorts to rolling-circle replication. T4 phage uses a different approach. Although it has a circular genetic
map it's DNA is linear dsDNA and remains so in the host and replicates to produce linear dsDNA copies with
'sticky ends' - single-stranded regions and concatemers are formed by enzymes joining together several
single copies of the genome by zipping together their single-stranded sticky ends. These are then cut-up
during packaging, as explained, with one phage head taking up slightly more than one complete genome's
worth and then the concatemer is cut and packaging moved on to the next phage head. This creates linear
dsDNA in which the two ends repeat and as there is some spare DNA this is called terminal redundancy.
This also creates cyclic permutations of the genetic code in different phages:
replicates by theta-replication, a form of bidirectional replication similar to that in bacteria like Escherichia
coli (see growth in bacteria). These initial copies ensure adequate production of viral products to initiate the
infection cycle, but later on, when assembling mature phages and packaging DNA into the phage heads it
resorts to rolling-circle replication. T4 phage uses a different approach. Although it has a circular genetic
map it's DNA is linear dsDNA and remains so in the host and replicates to produce linear dsDNA copies with
'sticky ends' - single-stranded regions and concatemers are formed by enzymes joining together several
single copies of the genome by zipping together their single-stranded sticky ends. These are then cut-up
during packaging, as explained, with one phage head taking up slightly more than one complete genome's
worth and then the concatemer is cut and packaging moved on to the next phage head. This creates linear
dsDNA in which the two ends repeat and as there is some spare DNA this is called terminal redundancy.
This also creates cyclic permutations of the genetic code in different phages:

Above: the structure of the T4 phage. The prolate head contains the
linear double-stranded DNa (dsDNA) of the viral genome. The six
long tail fibres adhere to specific targets on the host cell surface. The
six tail pins on the baseplate also assist adhesion to the host cell.
The tail tube connects the head to the baseplate and can contract
like a syringe, ejecting the tube in its core, acting like a hypodermic
needle, to inject the DNA into the target cell. The 6 whiskers (made of
a protein called fibritin) act as environmental sensors and can retract
the long tail fibres, switching the virus into a non-infectious state in
unfavourable conditions.
linear double-stranded DNa (dsDNA) of the viral genome. The six
long tail fibres adhere to specific targets on the host cell surface. The
six tail pins on the baseplate also assist adhesion to the host cell.
The tail tube connects the head to the baseplate and can contract
like a syringe, ejecting the tube in its core, acting like a hypodermic
needle, to inject the DNA into the target cell. The 6 whiskers (made of
a protein called fibritin) act as environmental sensors and can retract
the long tail fibres, switching the virus into a non-infectious state in
unfavourable conditions.
Herpesvirus has an icosahedral capsid (left) with 12
pentamers (pentagonal capsomeres or pentons) one at
each vertex and 150 hexameres (hexagonal capsomeres
or hexons) on the faces (60 hexons, 3 per face x 20 faces)
and edges (of which there are 30 each with 3 hexons) (T =
16). This capsid contains a toroidal (doughnut-shaped
ring) of DNA wound around a central protein cylinder. The
capsid is surrounded by a fibrous (in)tegument bounded
by a phospholiid bilayer envelope bearing viral spikes
(right, virus with section cut-away showing internal
structure).
Click images to enlarge.
pentamers (pentagonal capsomeres or pentons) one at
each vertex and 150 hexameres (hexagonal capsomeres
or hexons) on the faces (60 hexons, 3 per face x 20 faces)
and edges (of which there are 30 each with 3 hexons) (T =
16). This capsid contains a toroidal (doughnut-shaped
ring) of DNA wound around a central protein cylinder. The
capsid is surrounded by a fibrous (in)tegument bounded
by a phospholiid bilayer envelope bearing viral spikes
(right, virus with section cut-away showing internal
structure).
Click images to enlarge.
| Geminivirus consists of a double icosahedral capsid. |
| Parvoviruses are dodecahedral (like a 12-sided die). |
| The poliovirus capsid consists of a dodecahedron superimposed on an icosahedron. |
Listen to the virus song on Youtube!
Comment on this article!
| More detail on T4 bacteriophage More bacteriophages RNA viruses Adenovirus Poxviruses Ebolavirus Giruses - Giant Viruses Virus molecular architecture |